A Microbiology and Biotechnology Cross-Study: Reconstructing the Microbial Genome from Human Intestines
By Adi Permana
Editor Adi Permana

BANDUNG, itb.ac.id – The ITB SLST(School of Life Sciences and Technology) undergraduate program in Microbiology and master's program in Biotechnology held a combined class for the BM 3207 (Microbiomics) and BT6214 (Diet and Gut Microbiome) courses. Titled “Reconstruction of Ancient Microbial Genomes from the Human Gut”, the class invited Marsha Wibowo, Ph.D. from Harvard Medical School as the guest lecturer.
Microbiomes in humans consist of bacteria, archaea, viruses, and eukaryotes. “These microbes have the amazing potential to change our physiology in terms of health and disease,” defined Marsha. “Aside from optimizing our metabolism, they can protect our bodies against pathogens, allow our immune systems to work well, and directly or indirectly affect most of our physiological processes via basic functions.”
During her study at Harvard Medical School, Marsha researched factors that influence the gut microbiomes and metabolic health, ranging from lifestyle, diet, and body temperature. Lifestyle is classified into two types: industrial and non-industrial. While a non-industrial lifestyle follows the regular consumption of personally produced food without antibiotics, an industrial lifestyle refers to the sedentary habit of having antibiotic intake plus a western diet high in fats and carbohydrates.
“Both research and observation show that industrial lifestyle causes more chronic diseases and a significant decrease in the number of gut microbiomes,” Marsha added.
Nowadays, no humans can escape from the industrial lifestyle. Proof of the declining number of intestinal microbiomes is cited in Martin J. Blaser’s book “Missing Microbes”. “Blaser’s study in 2016 contains data that depicts the disappearance of these microbiomes in US citizens to be more rapid than other countries undergoing late urbanization,” Marsha continued.
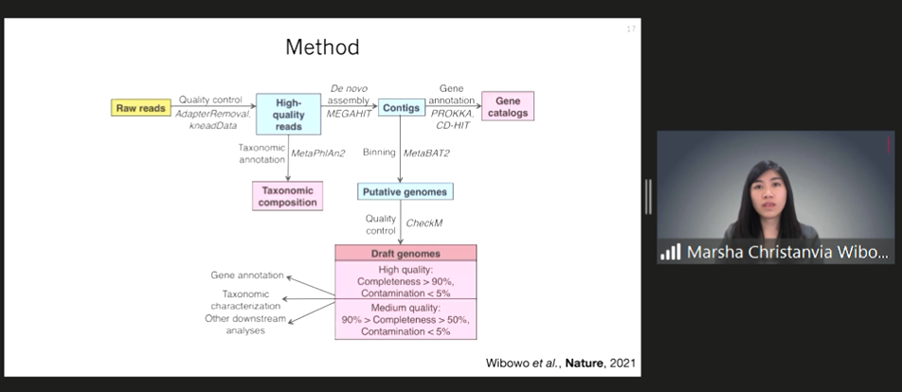
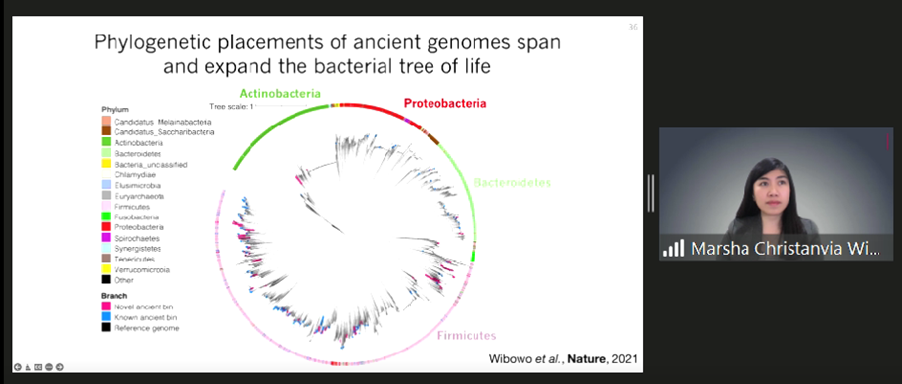
To examine the difference between the past and present levels of gut microbiomes in humans, Marsha assembled genomes of de novo microbes from palaeofaeces on a large scale. “From all eight authenticated human palaeofaeces samples aged 1000 to 2000 years originating at Southwestern USA and Mexico, we reconstructed 498 medium and high-quality microbial genomes,” Marsha disclosed. The results reveal that ancient genomes consist of many microbial phyla, including Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria.
This research showed that following the industrial lifestyle leads to less diverse gut microbiomes and more frequent incidents of chronic illnesses like obesity and autoimmune complications. Studying the intestinal microbiomes of human ancestors can provide insights into the symbiotic aspects of the changed microbiomes in today’s industrial world.
Reporter: Yoel Enrico Meiliano (Food Engineering, 2020)
Translator: Ruth Nathania (Environmental Engineering, 2019)



.jpg)

.jpg)


